|
|
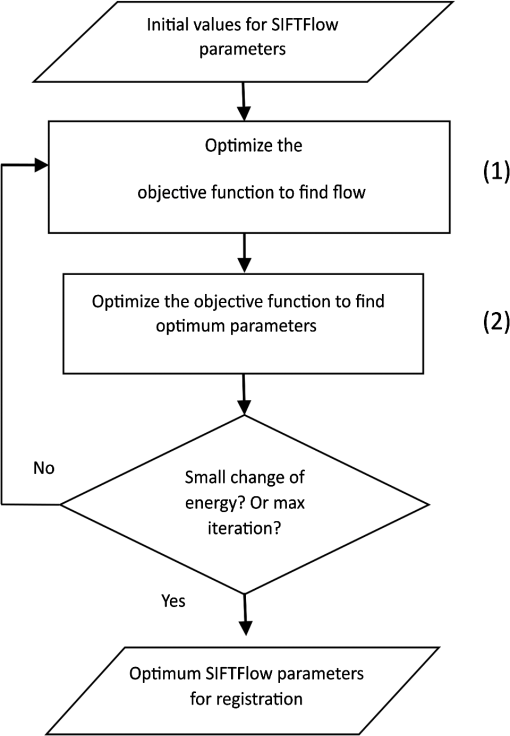
1.IntroductionBreast cancer is the most common cancer in women worldwide, the second leading cause of cancer death among women in developed countries following lung cancer, and the second most common cancer overall after skin cancer. The incidence rate of breast cancer is rising, and early diagnosis and accurate treatment planning play important roles in prolonging life expectancy with higher quality of life for patients. Medical imaging is the primary method for both detecting breast cancer and staging the disease, while pathology plays a vital role in diagnosis, prognosis, and prediction. The development of scanners that are capable of generating high-quality digital microscopy images of whole slides (WSIs) has led to increased research activity in the area of pathology image analysis. Although most research is focused on pathology as a two-dimensional (2-D) imaging modality, there is growing interest in exploring the additional information that can be obtained by reconstructing three-dimensional (3-D) histology images from stacks of 2-D WSIs. The 3-D reconstructions generated from histology images have two main applications. First, they can provide structural information at the microscopic resolution. For example, biologists have used this approach to study the morphology of embryonic development,1 the coronary system,2 and mammary glands3 in murine models, and 3-D histology reconstruction has also been applied to human breast cancer in the study of ductal carcinoma in situ (DCIS).4 A 3-D reconstruction also has the potential to improve our understanding of heterogeneity in tumors by allowing us to visualize the spatial distribution of different tumor markers. Second, 3-D histology volume reconstruction is an important intermediate step in the coregistration of WSI histology images to in vivo imaging techniques such as magnetic resonance imaging (MRI), optical coherence tomography (OCT), and ultrasound. Coregistration of medical images with histology provides a better understanding of how image appearances correlate with pathology and it allows in vivo imaging contrast mechanisms to be explored and validated using histology as the “gold standard.” In most cases, WSI histopathology images are obtained for standard glass microscope slides, and tissue specimens that are too large to fit on these slides must be cut into smaller blocks before sectioning. This means that regular WSIs cannot be used to generate 3-D volumes of larger clinical specimens such as whole prostate glands or breast lumpectomy specimens. Advances in digital scanner technology and specimen preparation and staining have made it possible for much larger slides of up to to be prepared and digitized in a process known as whole-mount histopathology.5 Whole-mount histopathology has been used to acquire cross-sectional images of complete breast6 and prostate7,8 specimens, and several semiautomated methods for reconstructing 3-D prostate histology volumes have been proposed.9–11 We are particularly interested in generating 3-D volumes from breast tumor specimens as it is known that 3-D features such as tumor margin, focality, and extent all have prognostic significance.5 This is, however, a significantly more challenging task than in the prostate for two main reasons. First, the breast tissue is composed largely of fat which means that the tissue sections are much more deformable and vulnerable to tearing. The second problem is that there are no clear boundaries to help guide the registration; this is especially true of lumpectomy specimens where the surgeon removes the tumor together with as little of the surrounding breast tissue as possible. Manual methods of reconstructing 3-D lumpectomy specimens have been described12,13 but they are very time-consuming and inaccurate. 1.1.Related WorkThe first step in generating a 3-D histology volume is to cut a series of very thin sections from tissue that has been embedded in a paraffin wax block. These sections are placed on glass slides, stained, and then scanned to generate digital WSIs. The placement of the tissue section onto the slide is not a precise process and the resulting image is affected by random in-plane translations and rotations, which have to be corrected for using rigid 2-D registration. In addition to this misalignment, folds, tears, and nonrigid deformations often occur during cutting, mounting, and staining the tissue sections and these must be corrected for using 2-D deformable registration. The final step in 3-D histology reconstruction involves stacking the 2-D images with the appropriate slice separation to form a volume and, in some cases, correcting for through-plane misalignment. Several approaches for correcting the 2-D misalignment and deformations have been described. Some groups have used extrinsic fiducial markers to align the slices,8,14 however, this is an invasive procedure, which has the potential to affect the pathologists’ ability to interpret the histological sections, and is frequently not permitted on clinical specimens. Both intensity- and landmark-based registration methods have the advantage that they do not interfere with the appearance of the tissue sections. Intensity-based methods have been shown to be very effective in medical image registration but they may fail in histology, where the intensities across histology images are not constant.15–17 Bağci et al.18,19 have demonstrated that intensity variations across images can lead to registration errors and this problem cannot be fixed through a simple intensity normalization approach. Feature-based image registration techniques20,21 rely on identifying corresponding features such as points and lines in both images, and the choice of a feature space plays a significant role in registration quality. Scale-invariant feature transform (SIFT) features22 take into account the spatial information obtained from the gradient of the pixel intensities and are invariant to brightness, rotation, and scale making them a suitable choice for use in histology. Many 3-D reconstruction methods rely solely on the information from the series of 2-D histology sections and either register slices one to another,15,23,24 or to a single reference section. 16,18,25 These methods assume that structures visible on one slice are still present in the adjacent slices but this may not be true if the separation between them is too great. Also local distortions of the histology sections, such as shrinkage, expansion, tears, and folds, which occur during the histological preparation procedures, reduce the similarities between consecutive sections and consequently make the alignment of the 2-D images much more challenging. To overcome this problem, some groups have made use of in vivo image, acquired before the tissue is removed, to constrain the 3-D reconstruction of the histology slices.10,11 This, however, is only possible if medical images are available and if it is possible to identify corresponding structures in both the histology and the in vivo images. In our previous work,3,26 we developed a pipeline for histology volume reconstruction, which used optical blockface images as reference images. Blockface images were obtained by photographing the surface of the tissue block before each section was cut, and the image of each histology section was then registered to its correspondent blockface image using 2-D rigid and deformable registration techniques. This is a practical solution when it is not feasible to densely sample the tissue using a small interslice separation and reference 3-D volumetric images of the specimen are not available. An edge-based registration method made use of points at the boundary between the tissue specimen and the surrounding paraffin block to carry out a rigid registration followed by a nonrigid deformation. We used implanted fiducial markers27 to measure the registration accuracy in mouse xenograft specimens and found that the deformable registration generated using the boundary points actually degraded the accuracy of the global rigid registration.26 This highlights the need to improve the deformable registration step in the pipeline. As mentioned earlier, the goal of our work is to generate 3-D histology volume reconstructions from whole-mount breast lumpectomy images, where the highly deformable nature of breast tissue and the lack of clear boundaries and anatomical structure make this a particularly difficult problem. Clarke et al.13 demonstrated that manually identified landmarks can be used to align a whole-mount lumpectomy histology image with a specimen cone-beam computed tomography slice and used this manual alignment to compare 3-D with 2-D tumor measurements,12 but this is a laborious and imprecise process. Booth et al.4 investigated the use of automated 3-D reconstruction of small breast tissue WSIs in the study of the biology of DCIS. They reconstructed the histology volume by selecting the middle section as the reference image and registering the neighboring sections to this reference image using the sum of squared differences of the neighboring pixel intensities.28 This registration proceeded from the center out with subsequent sections aligned to their neighbors and it relied on having sequential -thick sections making it impractical for use with whole-mount images. Reis et al.29 investigated the maximum spacing between breast histology sections required for 3-D histology reconstruction to a specific tolerance, focusing on the accuracy of reconstruction of individual ductal structures that were extracted from the WSI. They also selected the section at the center of the block as the reference image and used a pairwise rigid or affine registration approach to align the sections to the reference image. They concluded that a spacing of between sections was sufficient to assess ductal structures. However, in the clinical setting, the spacing between the breast sections is about 4 mm and methods that rely on slice-to-slice registration will likely fail. 1.2.Our ApproachOur goal, therefore, is to develop an automated method for generating 3-D histology volumes that can be applied to whole-mount breast lumpectomy or mastectomy specimens without manual intervention and without relying on closely spaced serial sections. This paper focuses on the first step in a 3-D volume reconstruction pipeline, which is the correction of deformations in the whole-mount images using a reference blockface image. In our previous work, we automatically identified points at the boundary between the tissue specimen and the surrounding paraffin block in both histology and blockface images and used these to generate a deformation field,26 but the results were disappointing. Here, we propose the use of an automatic feature-based image registration method, SIFTFlow, developed by Liu et al.,30,31 to align the whole-mount and blockface images. Using SIFT features which are invariant to brightness, rotation, and scale eliminates the shortcoming of the intensity-based methods, and by establishing a very dense set of corresponding points we hope to improve on our previous method which only used sparsely sampled edge points. We investigate the accuracy of the SIFTFlow algorithm as a function of the parameter settings used and then optimize these parameters for our specific application using fiducial markers implanted in a lumpectomy specimen to assess registration errors. Finally, we compare the performance of the proposed deformable registration method with the edge-based method described previously. Further processing steps to align the blockface images are required to complete the 3-D volume reconstruction once the deformations in the whole-mount images have been corrected. Large breast tissue specimens have to be cut into 4-mm-thick slices before paraffin embedding is carried out to facilitate tissue processing, which means that each whole-mount section is cut from a different block and additional tissue processing steps are required to realign the blockface images. We will report on the results of 3-D volume reconstruction in future work as a description of the full reconstruction pipeline is beyond the scope of this paper. 2.Materials and MethodsThis section describes the methods used to acquire whole-mount histology images and their corresponding blockface images and the pipeline for coregistration of these images using densely sampled SIFT features.22,30 We also propose an iterative method for optimizing the parameters used by the SIFTFlow algorithm. 2.1.Image DataTwo sets of image data were acquired which we will refer to as the training set and the test set. The training set of 10 whole-mount/blockface image pairs came from a single specimen and was used to determine an optimum set of parameters for the SIFTFlow algorithm and to carry out the comparison with the edge-based approach. The test dataset, comprising five image pairs from five different patients, was used to test the registration accuracy obtained using the optimized SIFTFlow method. For the training data, we were able to obtain Institutional Research Ethics Board approval to implant multimodality fiducial markers into part of a lumpectomy specimen from a patient with invasive ductal carcinoma (the rest of the specimen was retained in the anatomic pathology laboratory for routine clinical assessment). Four multimodality fiducial markers were implanted in the fresh tissue using a biopsy needle.27 The specimen was suspended in 3.5% agar gel and four additional fiducial markers were placed in the agar gel close to the specimen; these markers could not be used for the histology to blockface registration as the gel tended to become detached from the tissue during sectioning. The fresh tissue was sliced at 4 mm. Figure 1(a) shows one slice of the fresh tissue with the fiducial markers both in agar gel and the specimen. The tissue slices were then formalin fixed, processed, and embedded in paraffin6 to produce 10 blocks. One -thick whole mount section was obtained from each paraffin block. Fig. 1(a) 4-mm slice of the fresh tissue, (b) blockface image (90-deg angle), (c) blockface image (oblique angle), and (d) H&E-stained whole-mount image. 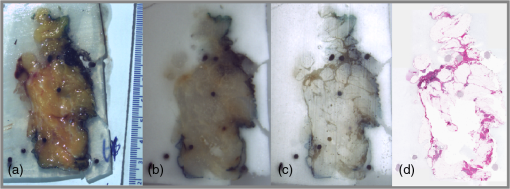 Immediately prior to cutting each section, a blockface image was captured using a telecentric lens. Imaging the blocks at a 90-deg angle provides an image with low contrast between the tissue at the surface of the block and the surrounding paraffin and also between different tissue types [Fig. 1(b)]. Positioning both the lens and light source at an oblique angle ( from 90-deg angle) produces a high-contrast image [Fig. 1(c)], but introduces a horizontal scaling artifact. We correct this by calculating a scaling factor based on the horizontal distances between the fiducial markers in the gel. After applying the scaling correction, the oblique blockface images are used as reference images. The pixel size of the blockface images was and the corrected blockface images were cropped to to remove excessive background. The tissue sections were stained with hematoxylin and eosin (H&E), which provides good contrast between fibroglandular tissue (purple), stroma (pink), and fatty tissue (mostly white), and is the most commonly used histological stain. They were then digitized at resolution, downsampled to the resolution of the blockface images, and padded with the average background pixel values to the size of blockface images [Fig. 1(d)]. For validation, a testing set of five pairs of blockface and histology images were also acquired from lumpectomy specimens from five different patients with the Institutional Research Ethics Board approval. It was not possible to get approval to implant fiducial markers in these specimens and only a single section was used from each specimen. A smaller lens was used to obtain the blockface images for these testing images, therefore, the resolution of blockface images and the downsampled whole-mount images was . 2.2.Registration Using Scale-Invariant Feature Transform FeaturesDense SIFT features22,31 are used, in this work, to find the rotation and translation of the histology images with respect to their correspondent blockface images. For each pixel, an eight-bin histogram of gradient orientations is calculated for 16 neighboring blocks, which results in features. Therefore, for each image, we have a 3-D SIFT image of dimension ().30 2.2.1.Affine registrationThe image pairs are first aligned using an affine registration. To find the rotation angle, shift parameters, and scaling factors, we minimized the Euclidean distance between the SIFT features of the histology and blockface images as follows: where and are the SIFT features of the blockface and the histology images, respectively, variable is the rotation angle, and are the shift values in the - and -directions, and and are the scale factors, which correct for any shrinkage and expansion of the tissue sections during processing. In the interest of speed, affine registration using fewer dense SIFT features was also investigated. Reducing the number of features to 10, i.e., , demonstrates results similar to those for . Therefore, the only first 10 features were used for rigid registration. Minimizing this objective function is an unconstrained multivariable optimization problem; therefore, the Nelder–Mead algorithm32 is used to find the registration parameters.2.2.2.Deformable registrationAfter the images have been roughly aligned using a similarity transformation, we use a deformable image registration to further improve the correspondence between the two images. Finding the pixel-wise displacement field is typically achieved by solving an optimization problem. Liu et al.30,31 showed that the SIFTFlow objective function works well for deformable registration of different scenes containing similar objects. They adopted a discrete, discontinuity preserving, flow estimation algorithm to match the dense SIFT features, instead of raw pixels. The SIFTFlow objective function [Eq. (2)] comprises three terms: a data term, a small displacement term, and a smoothness term where and are the dense SIFT features of the source and target images, and variable is the displacement of pixel . Parameter controls the displacement of the pixels and controls the smoothness of this displacement with respect to the neighboring pixels. Small values for these parameters allow for high deformations while higher values allow for milder deformations. The smoothness term, which encourages pairwise smoothness in the pixel displacements, is controlled by and is bounded by a threshold parameter . The use of a truncated L1-norm for the smoothness term accounts for outliers in the deformation field such as the presence of large tears, where neighboring pixels may be subjected to very different deformations.2.2.3.SIFTFlow parameter optimizationLiu et al.30,31 do not discuss their choice of regularization parameter values in their papers, suggesting that the default parameter settings of , , and used in their software implementation33 were suitable for the many image types used in Ref. 31 to illustrate the SIFTFlow method. Our task of registering breast histology images to blockface images, however, is very different from that of registering natural images due to the presence of tears, folds, and other artifacts in the histology slices and it is necessary to determine what effect the parameter settings have on registration accuracy and also to identify the most appropriate settings to use. We developed the iterative optimization method summarized in Fig. 2 to determine the most appropriate parameter settings for lumpectomy specimens. In the first step, the objective function [Eq. (2)] is minimized to find the displacement field using the initial parameter values from Ref. 33. In this step, the efficient belief propagation method31,34 is used to find the deformation field. In the second step, the objective function is optimized using the Nelder–Mead optimization algorithm to find updated parameter estimates for the displacement field obtained from the first step. The new estimated parameters are then used as inputs for the first step to find a new displacement field. These two steps iterate until either the change of the function value is lower than an upper limit or it reaches a maximum number of iterations. We carried out this parameter optimization on the training data set. Leave-one-out cross-validation (LOOCV) was adopted, i.e., the parameters were estimated using nine pairs of images and then tested on the image which was left out. The formulated objective function with LOOCV is shown as follows: The summation on is added for LOOCV.To make sure that the optimization was not influenced by the presence of the fiducial markers, the regions of the images with fiducial markers were marked manually for each pair of histology and blockface images after the rigid alignment of the images and then excluded from the calculation of the objective function. 2.3.Registration Using Edge PointsIn previous work, we used an edge-based registration method3,26 to align histology and blockface images from smaller murine tissue blocks. Here, we apply the same method to the whole-mount lumpectomy images to make a comparison with the SIFTFlow method. For the initial affine registration step, the segmented boundary points are aligned using Fourier descriptors35,36 and then the registration is refined using the iterative closest points37 algorithm. The aligned boundary points are then used as control points to find the nonrigid deformation, and a thin-plate spline is used to model the deformation field. The edge-based approach is simple to apply when there is a good contrast between the tissue and background in both histology and blockface images. In breast histology images, however, there is very low contrast between the fatty tissue and the background, which makes the automatic segmentation of the boundary points very challenging. Therefore, in this work, we manually segmented the boundaries of the tissue in both sets of images. 2.4.ValidationThe rigid alignment of the image pairs in the training set was assessed by measuring the mean target registration error (TRE) based on the center of the four fiducial markers implanted in the specimen. To measure the deformable error, we also selected some internal landmarks in each pair of images and calculated the mean TRE based on these internal points. We attempted to identify an equal number of evenly distributed landmarks in each image pair for both the training and the testing datasets. 3.Results3.1.Affine RegistrationRigid and affine registrations were carried out using the SIFT feature-based method described in Sec. 2.2.1 and the edge-based method described in Sec. 2.3. Figure 3 shows an example of one pair of blockface and histology images together with the aligned histology images obtained by using the edge- and the feature-based methods, respectively. The large amount of fatty tissue leads to very poorly defined tissue boundaries, particularly in the histology images, and it was necessary to manually define boundaries for the edge-based approach. Fig. 3Rigid registration of a histology image to the corresponding blockface image: (a) blockface image with boundary in blue (figure best viewed in color) and (b) histology image with manually defined boundary shown in red and blockface boundary in blue. Aligned histology images: (c) using edge-based method and (d) using dense SIFT features. The boundary of the blockface image is overlaid on both aligned histology images for comparison. 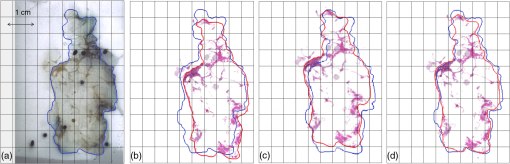 The TRE calculated using the center of the implanted fiducial markers in the tissue is reported in Table 1. The improvement in TRE when using dense SIFT feature-based method is small with respect to the edge-based method, but the feature-based method has the advantage that it is not dependent on image segmentation. The results in Table 1 also show that using a similarity transformation, which allows for scaling, improves the alignment compared with a simple rigid transformation. Table 1TREs for rigid and affine registration. Values are the mean TREs (± stdev) based on implanted fiducial markers.
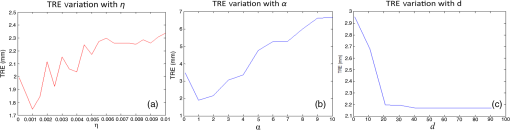
3.2.Deformable Registration3.2.1.Effect of regularization parameters on SIFTFlow registrationFigure 4 illustrates the effect of changing the parameters controlling the magnitude of the displacement of the pixels and the smoothness of this displacement . White areas on the images represent regions where compression has occurred. Fig. 4The deformation fields calculated with different settings for and are applied to a grid image to visualize the effects of changing the regularization parameters. Increasing results in smoother displacement: (a) , and (b) , . Increasing results in smaller displacements: (c) , and (d) , . Parameter was fixed at the default value of 40 in all cases. 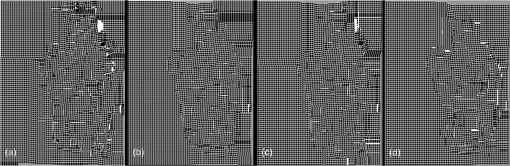 The effect of changing each of the regularization parameters was assessed by holding two of the parameters fixed at the default values and then varying the third parameter. The plots in Fig. 5 show the mean TRE calculated using the fiducial markers in all 10 training image pairs as each parameter is varied over a range of values. We see that the largest variation is caused by changes in , the smoothness parameter, and Fig. 5(c) suggests that changing has very little effect once . We, therefore, kept fixed at the default value of 40 when carrying out the multivariate optimization outlined in Sec. 2.2.3. 3.2.2.Identifying paired landmarks for the assessment of deformable registrationMost of the landmarks were selected at the corners of the fibroglandular tissue and a few at the cross section of the blood vessels, where the correspondence of the points in histology and blockface images could be determined. It was not possible to identify landmarks in the fatty tissue with confidence. Figure 6 shows two pairs of blockface and histology images. The smaller section contains mostly fat, while the larger section has more fibroglandular tissue making it possible for more landmarks to be identified. The number of manually selected landmarks varied for different sections in the training dataset as there was a wide variation in section size and composition and, although we attempted to evenly distribute landmarks throughout the images, this was not always possible as can be seen in Fig. 6. The number of landmarks in the 10 training sections varied from 4 to 10. Fig. 6Two pairs of blockface and histology images with different sizes and composition. Top row shows the (a) blockface and (b) histology images of a small section, which mostly contains fat. Bottom row shows the (c) blockface and (d) histology images of a larger section with more fibroglandular tissue and higher number of selected landmarks. The arrows on the blockface images show the selected landmarks. 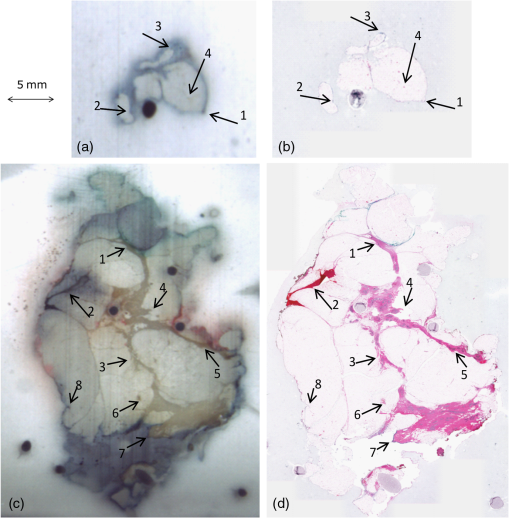 The five sections taken from five different patients were all selected through the center of the specimens and as well as being larger, they also contained more fibroglandular tissue allowing us to identify 10 landmarks in each test image pair. 3.2.3.Comparing edge-based deformable registration with SIFTFlowThe procedure outlined in Fig. 2 was used to estimate the optimum parameters and in the SIFTFlow objective function ( was fixed at 40 throughout). The optimized parameters and the associated mean TREs (calculated using both the implanted fiducials and the manually defined landmarks) from the LOOCV experiment using 10 pairs of images are reported in Table 2. Table 2Average TREs for LOOCV experiment using edge-based SIFTFlow with default parameters, and SIFTFlow with optimized parameters measured using both fiducial markers and manually selected landmarks. Values are given as mean (stdev) in mm.
In case no. 02, the error is very high; this is because a part of the tissue section is missing on the histology slide leading to a large discordance between the stained section and the corresponding blockface image. For the TREs measured using fiducial markers, the results confirm that optimizing the regularization parameters significantly improves registration accuracy ( using a paired two tailed -test, normal distribution of TREs was confirmed using a Kolmogorov–Smirnov test). The average difference between the TRE obtained from the optimized and default parameters is 0.67 mm, which corresponds to at resolution. The results of the deformable registration using the edge-based method are also shown in Table 2. Errors were significantly higher than those obtained with the SIFTFlow method. Figure 7 shows one of the blockface images and the corresponding histology image after registration with each of the methods tested. The circle in Figs. 7(a) and 7(b) highlights an area of tissue that is missing on the histology slides. In Fig. 7(b), a part of the tissue is missing and the segmented tissue boundaries are different in the histology and blockface images, resulting in poorer performance of the edge-based registration method compared to the feature-based method. It has also been noticed that with the edge-based method, as expected, the structures closer to the edge of the tissue are generally better aligned than the structures further away from the boundary points. In Figs. 7(c) and 7(d), arrows point out areas of difference in the default and optimized SIFTFlow results. Fig. 7Images from case no. 04: (a) blockface image and (b) histology registered using edge-based method; the circle shows a missing part of the tissue section in histology image, which can degrade the result of the registration. c) Histology registered using default SIFTFlow parameters and (d) histology registered using optimized SIFTFlow. The arrows point to some of the areas that are more accurately registered when the optimized parameters are used. 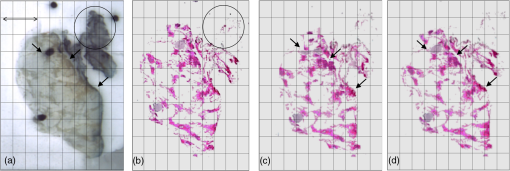 To obtain a single set of SIFTFlow parameters to use in the test dataset, we repeated the optimization procedure using all 10 image pairs and obtained values of and . 3.2.4.Accuracy of optimized SIFTFlow on an independent test datasetTo confirm that parameters optimized using the training dataset are suitable for different breast specimens, we applied both these parameters and the default parameters to five pairs of blockface and histology images from five different patients and the results are presented in Table 3. The edge-based method was not applied to this data as the results from the training data clearly indicated that it is inferior to the SIFTFlow method. Table 3TRE based on the internal structures of five histology sections from five different patients with the default parameters (η=0.005) and α=2.00) and the optimized parameters (η=4.9×10−3 and α=1.169).
TREs were comparable with those obtained in the training set and the optimized parameters resulted in an improvement over the default parameters. The mean registration error was calculated as for this independent test set. 4.DiscussionIn this work, we have demonstrated that SIFTFlow is successful in aligning blockface and histology images and we have developed a method of tuning the regularization parameters used by this approach. SIFTFlow removes the need for image segmentation and is more accurate than our previously described edge-based approach. Another advantage is that it uses SIFT features, which are relatively robust to variations in illumination and intensity in different images. This means the same technique could be used for registration of histology with other imaging modalities such as ex vivo MRI or OCT or for the alignment of histology images with different complementary stains. For the rigid registration step, we found that it was useful to use a similarity transformation model rather than a rigid transformation suggesting that the tissue sections may expand when they are placed in a water bath during the mounting procedure. We also found that a lower value of than that suggested by Liu et al. significantly improved the results of registration. The mixture of highly deformable fatty tissue and less deformable fibroglandular tissue in breast tissue specimens, together with the presence of folds and tears which inevitably occur when mounting a very flexible tissue section onto a glass slide, leads to the presence of more complex deformation fields in histology images than would have been present in the natural images used to demonstrate the SIFTFlow method in Ref. 31. Lower values for parameter have the effect of relaxing the smoothness constraint in the objective function and this allows for higher variability in the deformation field. Our method for carrying out a joint optimization of the regularization parameters allows SIFTFlow to be tuned for specific tissue types which could have very different biomechanical properties leading to differences in the range of deformations. One of the limiting factors of the proposed parameter estimation approach for SIFTFlow is that it is sensitive to the initial values. In this work, we used the parameters published by Liu et al. as initial values for optimization. Currently, we are working on a preoptimization fast search algorithm to find suitable ranges for initial values to overcome this problem. The motivation of this work was to correct for deformations in the histology images prior to reconstructing a 3-D volume. We make the assumption that the blockface images are free from distortion and can, therefore, be used as a reference image. In practice, significant distortions occur as a result of the tissue processing steps carried out to embed the tissue block in paraffin and these will also have to be accounted for if the 3-D volume is to be subsequently aligned with 3-D ultrasound or MRI images. Although our results are encouraging, in applications where a region of interest in an aligned histology image needs to be assessed at a higher resolution, even a submillimeter misalignment could become very significant. For example, when a region of interest in a medical image is correlated with the correspondent region in a histology image, the assessment of the region at higher resolution is required to reveal the underlying structures in the histology image. An average difference of 0.67 mm corresponds to an error of 4 pixels at resolution and 335 pixels at resolution, which means that zooming in a region of interest selected at low resolution could lead to the selection of a totally misaligned patch at a higher resolution. The accuracy of the image registration will be further improved by the employment of multiscale image registration methods in future work. 5.ConclusionIn this paper, we have presented an automatic approach for correcting for deformations in whole-mount histology images from human breast lumpectomy specimens. The contribution of the paper is twofold: (1) we have demonstrated that a noninvasive, automatic, and feature-based image registration method can be applied to the task of aligning histology images with optical blockface images and (2) we have proposed a method for optimizing this method and shown that this significantly reduced TRE. Quantitative and qualitative evaluations of the image registration results for different breast specimens demonstrate lower registration error when optimized parameters are used. The optimized feature-based method also outperforms a previously described edge-based method. This method will be used as a preprocessing step in the generation of 3-D volume reconstructions of whole-mount histology images. AcknowledgmentsThe authors would like to thank the Biomarker Imaging Research Laboratory at Sunnybrook Research Institute for preparing the specimens and acquiring the whole-mount and blockface images. Support for this work was provided by Ontario Institute for Cancer Research and the National Institute of Health (No. 1U24CA199374-01). ReferencesR. M. Brune et al.,
“A three-dimensional model of the mouse at embryonic day 9,”
Dev. Biol., 216
(2), 457
–468
(1999). http://dx.doi.org/10.1006/dbio.1999.9500 DEBIAO 0012-1606 Google Scholar
L. Fónyad et al.,
“3-dimensional digital reconstruction of the murine coronary system for the evauation of chronic allograft vasculopathy,”
Diagn. Pathol., 10
(1), 16
(2015). http://dx.doi.org/10.1186/s13000-015-0248-6 DMPAES 1052-9551 Google Scholar
R. Shojaii et al.,
“Reconstruction of 3-dimensional histology volume and its application to study mouse mammary glands,”
J. Vis. Exp., 89 e51325
(2014). http://dx.doi.org/10.3791/51325 Google Scholar
M. E. Booth et al.,
“Three-dimensional reconstruction of ductal carcinoma in situ with virtual slides,”
Histopathology, 66
(7), 966
–973
(2015). http://dx.doi.org/10.1111/his.12561 HISTDD 1365-2559 Google Scholar
G. M. Clarke et al.,
“Increasing specimen coverage using digital whole-mount breast pathology: implementation, clinical feasibility and application in research,”
Comput. Med. Imaging Graphics, 35
(7–8), 531
–541
(2011). http://dx.doi.org/10.1016/j.compmedimag.2011.05.002 Google Scholar
L. Sun et al.,
“An improved processing method for breast whole-mount serial sections for three-dimensional histopathology imaging,”
Am. J. Clin. Pathol., 131
(3), 383
–392
(2009). http://dx.doi.org/10.1309/AJCPVBZZ4IKJHY3U AJCPAI 0002-9173 Google Scholar
A. Boyes et al.,
“Prostate tissue analysis immediately following magnetic resonance imaging guided transurethral ultrasound thermal therapy,”
J. Urol., 178
(3), 1080
–1085
(2007). http://dx.doi.org/10.1016/j.juro.2007.05.011 Google Scholar
E. Gibson et al.,
“3D prostate histology reconstruction: an evaluation of image-based and fiducial-based algorithms,”
Med. Phys., 40
(9), 093501
(2013). http://dx.doi.org/10.1118/1.4816946 MPHYA6 0094-2405 Google Scholar
A. D. Ward et al.,
“Prostate: registration of digital histopathologic images to in vivo MR images acquired by using endorectal receive coil,”
Radiology, 263
(3), 856
–864
(2012). http://dx.doi.org/10.1148/radiol.12102294 RADLAX 0033-8419 Google Scholar
G. Nir et al.,
“Registration of whole-mount histology and volumetric imaging of the prostate using particle filtering,”
IEEE Trans. Med. Imaging, 33
(8), 1601
–1613
(2014). http://dx.doi.org/10.1109/TMI.2014.2319231 ITMID4 0278-0062 Google Scholar
G. Xiao et al.,
“Determining histology-MRI slice correspondences for defining MRI-based disease signatures of prostate cancer,”
Comput. Med. Imaging Graphics, 35
(7–8), 568
–578
(2011). http://dx.doi.org/10.1016/j.compmedimag.2010.12.003 Google Scholar
G. M. Clarke et al.,
“3D pathology volumetric technique: a method for calculating breast tumour volume from whole-mount serial section images,”
Int. J. Breast Cancer, 2012 691205
(2012). http://dx.doi.org/10.1155/2012/691205 Google Scholar
G. M. Clarke et al.,
“Validation of cone-beam CT measurements of tumour burden using three-dimensional histopathology: initial results for a lumpectomy,”
Lect. Notes Comput. Sci., 5116 440
–446
(2008). http://dx.doi.org/10.1007/978-3-540-70538-3_61 LNCSD9 0302-9743 Google Scholar
M. S. Breen, R. S. Lazebnik and D. L. Wilson,
“Three-dimensional registration of magnetic resonance image data to histological sections with model-based evaluation,”
Ann. Biomed. Eng., 33
(8), 1100
–1112
(2005). http://dx.doi.org/10.1007/s10439-005-5778-8 ABMECF 0090-6964 Google Scholar
T. Ju et al.,
“3D volume reconstruction of a mouse brain from histological sections using warp filtering,”
J. Neurosci. Methods, 156
(1–2), 84
–100
(2006). http://dx.doi.org/10.1016/j.jneumeth.2006.02.020 JNMEDT 0165-0270 Google Scholar
G. Malandain et al.,
“Fusion of autoradiographs with an MR volume using 2-D and 3-D linear transformations,”
NeuroImage, 23
(1), 111
–127
(2004). http://dx.doi.org/10.1016/j.neuroimage.2004.04.038 NEIMEF 1053-8119 Google Scholar
S. Ourselin et al.,
“Automatic alignment of histological sections for 3D reconstruction and analysis,”
(1998). Google Scholar
U. Bağci and L. Bai,
“Registration of standardized histological images in feature space,”
Proc. SPIE, 6914 69142V
(2008). http://dx.doi.org/10.1117/12.770219 PSISDG 0277-786X Google Scholar
U. Bağci, J. K. Udupa and L. Bai,
“The role of intensity standardization in medical image registration,”
Pattern Recognit. Lett., 31
(4), 315
–323
(2010). http://dx.doi.org/10.1016/j.patrec.2009.09.010 PRLEDG 0167-8655 Google Scholar
E. Guest et al.,
“Robust point correspondence applied to two- and three-dimensional image registration,”
IEEE Trans. Pattern Anal. Mach. Intell., 23
(2), 165
–179
(2001). http://dx.doi.org/10.1109/34.908967 ITPIDJ 0162-8828 Google Scholar
F. S. Cohen et al.,
“Automatic matching of homologous histological sections,”
IEEE Trans. Biomed. Eng., 45 642
–649
(1998). http://dx.doi.org/10.1109/10.668755 IEBEAX 0018-9294 Google Scholar
D. G. Lowe,
“Distinctive image features from scale-invariant keypoints,”
Int. J. Comput. Vision, 60
(2), 91
–110
(2004). http://dx.doi.org/10.1023/B:VISI.0000029664.99615.94 IJCVEQ 0920-5691 Google Scholar
E. Guest and R. Baldock,
“Automatic reconstruction of serial sections using the finite element method,”
Bioimaging, 3
(4), 154
–167
(1995). http://dx.doi.org/10.1002/1361-6374(199512)3:4<154::AID-BIO2>3.0.CO;2-M BOIMEL 0966-9051 Google Scholar
S. Wirtz et al.,
“Super-fast elastic registration of histologic images of a whole rat brain for three-dimensional reconstruction,”
Proc. SPIE, 5370 328
–334
(2004). http://dx.doi.org/10.1117/12.534110 PSISDG 0277-786X Google Scholar
U. Bağci and L. Bai,
“Automatic best reference slice selection for smooth volume reconstruction of a mouse brain from histological images,”
IEEE Trans. Med. Imaging, 29
(9), 1688
–1696
(2010). http://dx.doi.org/10.1109/TMI.2010.2050594 ITMID4 0278-0062 Google Scholar
R. Shojaii et al.,
“Validation of histology image registration,”
Proc. SPIE, 7962 79621E
(2011). http://dx.doi.org/10.1117/12.878762 Google Scholar
R. Shojaii and A. Martel,
“Multi-modality fiducial marker for validation of registration of medical images with histology,”
Proc. SPIE, 7623 762331
(2010). http://dx.doi.org/10.1117/12.845551 Google Scholar
N. Roberts et al.,
“Toward routine use of 3D histopathology as a research tool,”
Am. J. Pathol., 180
(5), 1835
–1842
(2012). http://dx.doi.org/10.1016/j.ajpath.2012.01.033 AJPAA4 0002-9440 Google Scholar
S. Reis et al.,
“Minimum slice spacing required to reconstruct 3D shape for serial sections of breast tissue for comparison with medical imaging,”
Proc. SPIE, 9420 94200K
(2015). http://dx.doi.org/10.1117/12.2081909 Google Scholar
C. Liu et al.,
“SIFT flow: dense correspondence across different scenes,”
Eur. Conf. Comput. Vision, 1
(1), 28
–42
(2008). http://dx.doi.org/10.1007/978-3-540-88690-7_3 Google Scholar
C. Liu, J. Yuen and A. Torralba,
“SIFT flow: dense correspondence across scenes and its applications,”
IEEE Trans. Pattern Anal. Mach. Intell., 33
(5), 978
–994
(2011). http://dx.doi.org/10.1109/TPAMI.2010.147 ITPIDJ 0162-8828 Google Scholar
B. J. Nelder and R. Mead,
“A simplex method for function minimization,”
Comput. J., 7 308
–313
(1965). http://dx.doi.org/10.1093/comjnl/7.4.308 Google Scholar
C. Liu, J. Yuen and A. Torralba,
“SIFT flow: Matlab/C++ code,”
(2011) http://people.csail.mit.edu/celiu/SIFTflow/SIFTflow.zip September 2016). Google Scholar
P. F. Felzenszwalb and D. P. Huttenlocher,
“Efficient belief propagation for early vision,”
Int. J. Comput. Vision, 70 41
(2006). http://dx.doi.org/10.1007/s11263-006-7899-4 IJCVEQ 0920-5691 Google Scholar
F. Kuhl and C. Giardina,
“Elliptic Fourier features of a closed contour,”
Comput. Graphics Image Process., 18 236
–258
(1982). http://dx.doi.org/10.1016/0146-664X(82)90034-X Google Scholar
W. Duan et al.,
“Automatic object and image alignment using Fourier descriptors,”
Image, 26 1196
–1206
(2008). http://dx.doi.org/10.1016/j.imavis.2008.01.009 IMGSBL Google Scholar
P. Besl and N. McKay,
“A method for registration of 3-D shapes,”
IEEE Trans. Pattern Anal. Mach. Intell., 14
(2), 239
–256
(1992). http://dx.doi.org/10.1109/34.121791 ITPIDJ 0162-8828 Google Scholar
BiographyRushin Shojaii is a PhD candidate in the department of medical biophysics at the University of Toronto. She received her BSc and MSc degrees in electrical and computer engineering. Her main areas of interest are medical image registration and coregistration of histology images with medical images. Anne L. Martel is a senior scientist at Sunnybrook Research Institute and an associate professor in the Department of Medical Biophysics, University of Toronto. She is a senior member of SPIE. Her research focuses on the analysis of medical images and computer-aided diagnosis. She is the cofounder of Pathcore Inc. (Toronto, Canada), a digital pathology company that develops and maintains the freely available Sedeen Viewer. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||